Note
Go to the end to download the full example code.
Create a full-heart model#
This example shows how to process a case from Rodero et al. (2021) into a simulation-ready heart model.
Perform the required imports#
Import the required modules and set relevant paths, including that of the working directory and generated model.
import json
import os
from pathlib import Path
import ansys.health.heart.models as models
from ansys.health.heart.pre.database_utils import get_compatible_input
from ansys.health.heart.utils.download import download_case_from_zenodo, unpack_case
# specify a download directory
download_folder = Path.home() / "pyansys-heart" / "downloads"
# Download a compatible case from the Zenodo database.
tar_file = download_case_from_zenodo("Rodero2021", 1, download_folder, overwrite=False)
# Unpack the case to get the input CASE or VTK file.
case_file = unpack_case(tar_file)
# Specify the working directory. This code uses the directory of the CASE file.
workdir = os.path.join(os.path.dirname(case_file), "FullHeart")
if not os.path.isdir(workdir):
os.makedirs(workdir)
# Specify paths to the model, input, and part definitions.
path_to_model = os.path.join(workdir, "heart_model.vtu")
path_to_input = os.path.join(workdir, "input_model.vtp")
path_to_part_definitions = os.path.join(workdir, "part_definitions.json")
Note
You can also manually download the CASE or VTK files from the Strocchi 2020 and Rodero 2021 databases. For more information, see:
Alternatively, you can simply click one of the buttons at the bottom of this page to download a CASE file for the Rodero 2021 database in an IPYNB, PY, or ZIP format.
Convert the VTK file to a compatible input format#
input_geom, part_definitions = get_compatible_input(
case_file, model_type="FullHeart", database="Rodero2021"
)
# Note that the input model and part definitions can be saved for later use.
# Save input geometry and part definitions.
input_geom.save(path_to_input)
with open(path_to_part_definitions, "w") as f:
json.dump(part_definitions, f, indent=True)
Create a heart model#
Create a full-heart model.
model = models.FullHeart(working_directory=workdir)
# Load input model generated in an earlier step.
model.load_input(input_geom, part_definitions, "surface-id")
# Mesh the volume of all structural parts.
model.mesh_volume(use_wrapper=True, global_mesh_size=2.0, _global_wrap_size=2.0)
# Update the model and extract the required anatomical features.
model.update()
# Optionally save the simulation mesh as a VTK object for "offline" inspection.
model.mesh.save(os.path.join(model.workdir, "simulation-mesh.vtu"))
model.save_model(os.path.join(model.workdir, "heart_model.vtu"))
# Print some information about the processed model.
print(model)
# Print part names.
print(model.part_names)
GENERAL:
total_num_tets: 332383
total_num_nodes: 72347
PARTS:
Left ventricle:
num_tets: 141000
SURFACES:
Left ventricle endocardium:
num_faces: 7421
Left ventricle epicardium:
num_faces: 8123
Left ventricle septum:
num_faces: 0
CAPS:
mitral-valve:
num_nodes: 67
aortic-valve:
num_nodes: 44
Right ventricle:
num_tets: 67893
SURFACES:
Right ventricle endocardium:
num_faces: 8875
Right ventricle epicardium:
num_faces: 9324
Right ventricle septum:
num_faces: 2758
CAPS:
tricuspid-valve:
num_nodes: 93
pulmonary-valve:
num_nodes: 58
Septum:
num_tets: 42734
SURFACES: {}
CAPS: {}
Left atrium:
num_tets: 31372
SURFACES:
Left atrium endocardium:
num_faces: 5740
Left atrium epicardium:
num_faces: 4369
CAPS:
mitral-valve-atrium:
num_nodes: 45
right-superior-pulmonary-vein:
num_nodes: 37
left-superior-pulmonary-vein:
num_nodes: 35
right-inferior-pulmonary-vein:
num_nodes: 31
left-atrium-appendage:
num_nodes: 28
left-inferior-pulmonary-vein:
num_nodes: 26
Right atrium:
num_tets: 32288
SURFACES:
Right atrium endocardium:
num_faces: 7168
Right atrium epicardium:
num_faces: 5564
CAPS:
tricuspid-valve-atrium:
num_nodes: 73
inferior-vena-cava:
num_nodes: 35
superior-vena-cava:
num_nodes: 34
Aorta:
num_tets: 12780
SURFACES:
Aorta wall:
num_faces: 4290
CAPS: {}
Pulmonary artery:
num_tets: 4316
SURFACES:
Pulmonary artery wall:
num_faces: 1348
CAPS: {}
CAVITIES:
Left ventricle cavity:
volume: 120446.96049660804
Right ventricle cavity:
volume: 185648.59541736593
Left atrium cavity:
volume: 68001.90309648328
Right atrium cavity:
volume: 110948.18865153291
['Left ventricle', 'Right ventricle', 'Septum', 'Left atrium', 'Right atrium', 'Aorta', 'Pulmonary artery']
Visualize results#
Visualize and inspect the components of the model by accessing various properties or attributes and invoking methods.
print(f"Volume of LV cavity: {model.left_ventricle.cavity.volume} mm^3")
print(f"Volume of LV cavity: {model.left_atrium.cavity.volume} mm^3")
# Plot the remeshed model.
model.plot_mesh(show_edges=False)
# Plot the endocardial surface of the left ventricle.
model.left_ventricle.endocardium.plot(show_edges=True, color="r")
# Loop over all cavities and plot them in a single window with PyVista.
import pyvista as pv
cavities = pv.PolyData()
for c in model.cavities:
cavities += c.surface
cavities.plot(show_edges=True)
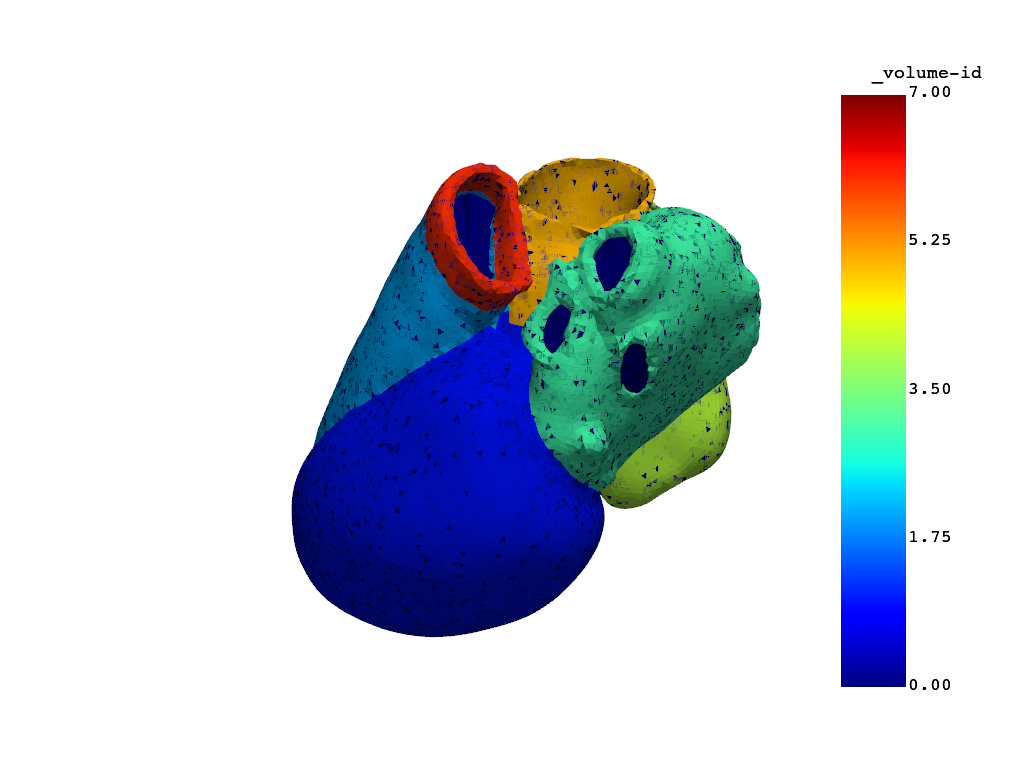

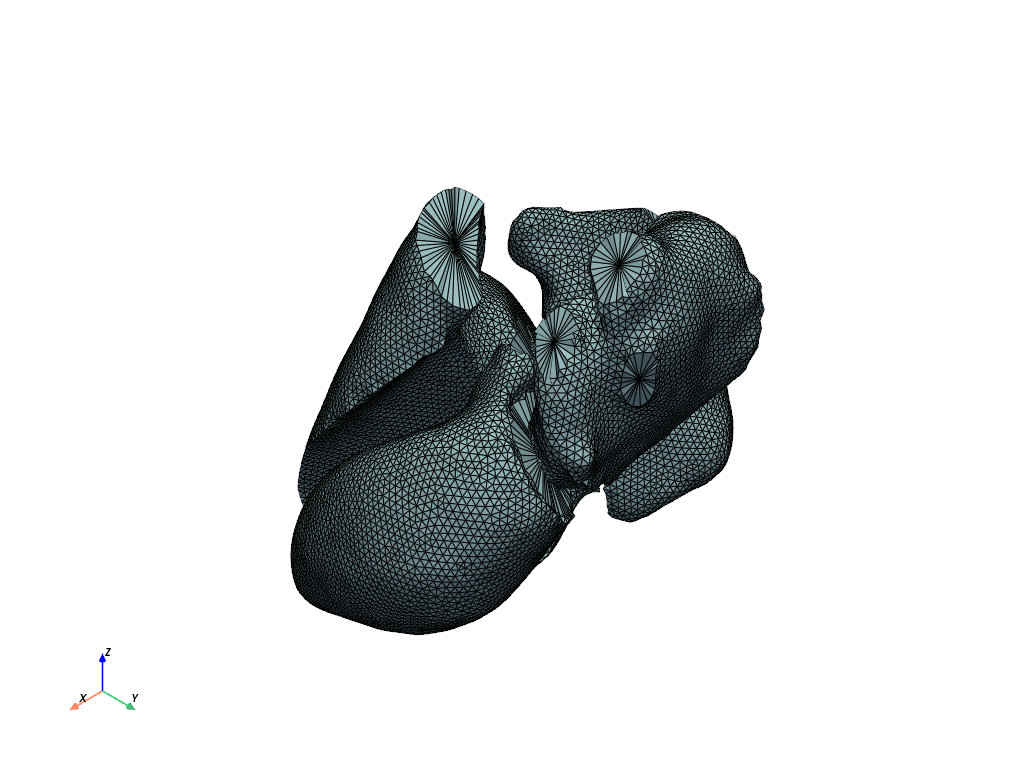
Volume of LV cavity: 120446.96049660804 mm^3
Volume of LV cavity: 68001.90309648328 mm^3
Total running time of the script: (7 minutes 44.941 seconds)

