Note
Go to the end to download the full example code.
Generate atrial fibers#
This example shows how to generate atrial fibers using the Laplace-Dirichlet Rule-Based (LDRB) method.
Warning
When using a standalone version of the DPF Server, you must accept the license terms. To accept these terms, you can set this environment variable:
import os
os.environ["ANSYS_DPF_ACCEPT_LA"] = "Y"
Perform the required imports#
Import the required modules and set relevant paths, including that of the working directory, model, and LS-DYNA executable file. This example uses DEV-104373-g6d20c20aee.
import os
from pathlib import Path
import numpy as np
import pyvista as pv
from ansys.health.heart.examples import get_preprocessed_fullheart
import ansys.health.heart.models as models
from ansys.health.heart.simulator import BaseSimulator, DynaSettings
# Specify the path to the working directory and heart model. The following path assumes
# that a preprocessed model is already available
workdir = Path.home() / "pyansys-heart" / "downloads" / "Rodero2021" / "01" / "FullHeart"
path_to_model, path_to_partinfo, _ = get_preprocessed_fullheart()
# Specify LS-DYNA path
lsdyna_path = r"ls-dyna_smp"
# Load heart model
model: models.FourChamber = models.FourChamber.load_model(
path_to_model, path_to_partinfo, working_directory=workdir
)
Instantiate the simulator#
Instantiate the simulator and modify options as needed.
Note
The DynaSettings object supports several LS-DYNA versions and platforms,
including smp, intempi, msmpi, windows, linux, and wsl.
Choose the one that works for your setup.
# instantiate LS-DYNA settings of choice
dyna_settings = DynaSettings(
lsdyna_path=lsdyna_path, dynatype="intelmpi", num_cpus=4, platform="windows"
)
simulator = BaseSimulator(
model=model,
dyna_settings=dyna_settings,
simulation_directory=os.path.join(workdir, "simulation"),
)
simulator.settings.load_defaults()
# remove fiber/sheet information if it already exists
model.mesh.cell_data["fiber"] = np.zeros((model.mesh.n_cells, 3))
model.mesh.cell_data["sheet"] = np.zeros((model.mesh.n_cells, 3))
Compute atrial fibers#
# compute left atrium fiber
la = simulator.compute_left_atrial_fiber()
# Import the appendage landmarks.
from ansys.health.heart.pre.database_utils import right_atrium_appendage_landmarks
# Get the right atrium appendage landmark of the first case of Rodero2021.
right_atrium_appendage_coordinates = right_atrium_appendage_landmarks.get("Rodero2021").get(1)
# Appendage apex point should be manually given to compute right atrium fiber
ra = simulator.compute_right_atrial_fiber(appendage=right_atrium_appendage_coordinates)
Note
You might need to define an appropriate point for the right atrial appendage. The list specifies the x, y, and z coordinates close to the appendage.
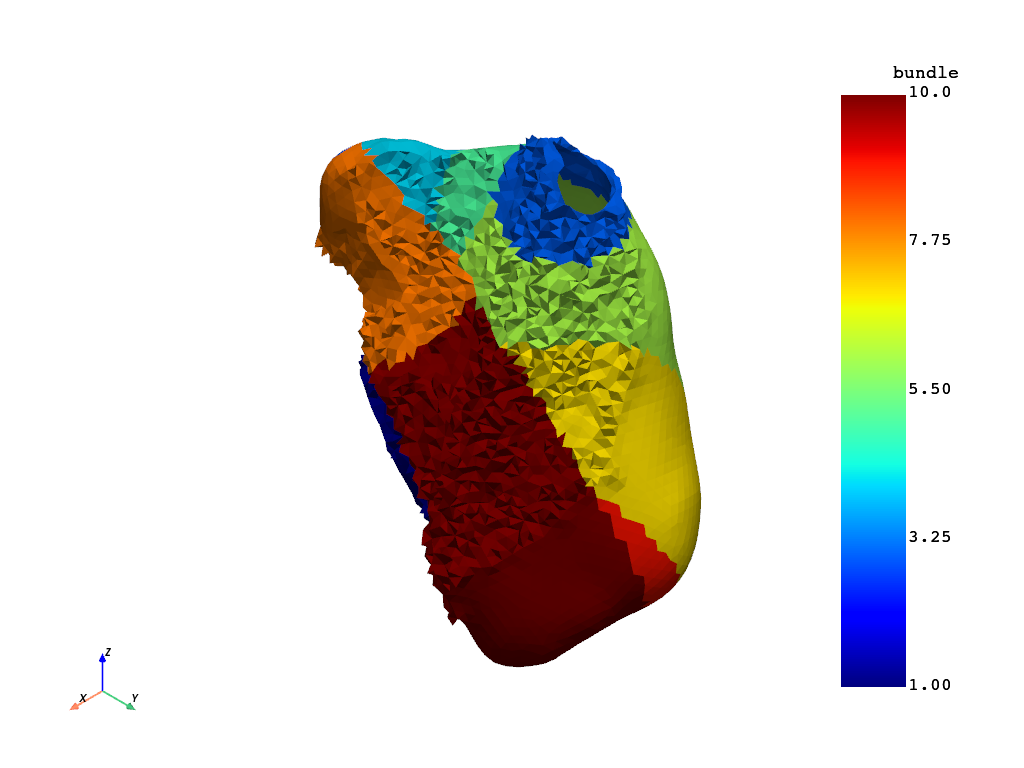
Plot left atrial bundles#
la.cell_data["bundle"] = la.cell_data["bundle"].astype(np.int32)
la.set_active_scalars("bundle")
la.plot()

Plot right atrial bundles#
ra.cell_data["bundle"] = ra.cell_data["bundle"].astype(np.int32)
ra.set_active_scalars("bundle")
ra.plot()
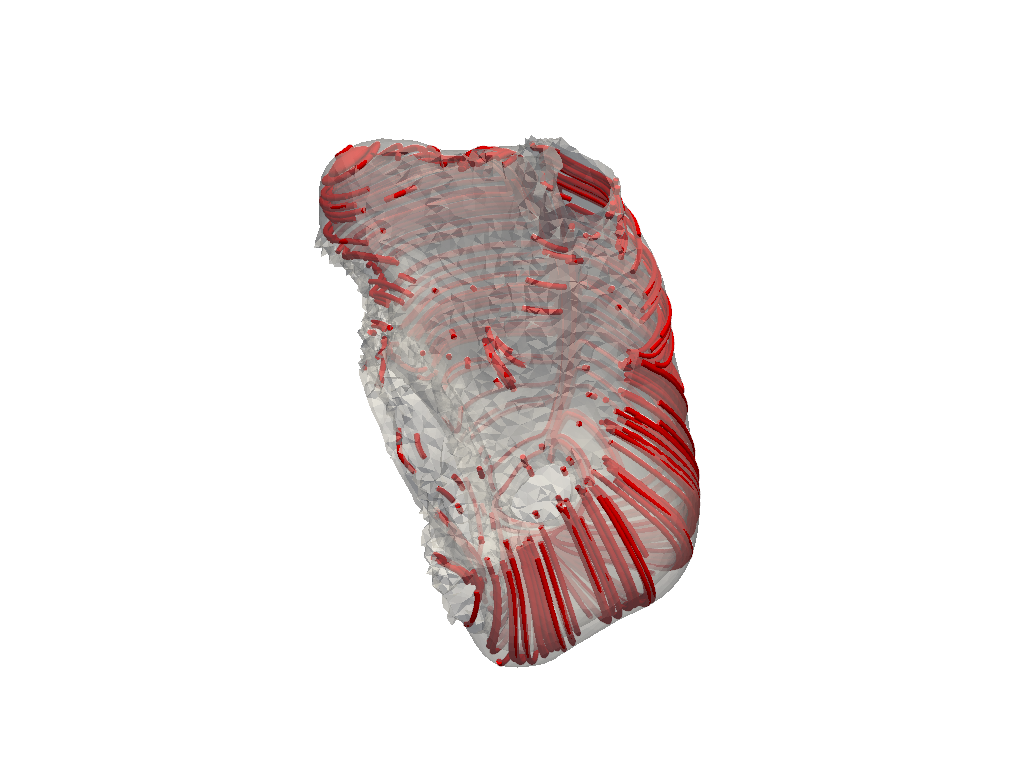
Plot left atrial fibers#
# plot left atrial fibers
plotter = pv.Plotter()
mesh = la.ctp()
streamlines = mesh.streamlines(vectors="e_l", source_radius=50, n_points=5000)
tubes = streamlines.tube()
plotter.add_mesh(mesh, opacity=0.5, color="white")
plotter.add_mesh(tubes, color="red")
plotter.show()

Plot right atrial fibers#
# plot right atrial fibers
plotter = pv.Plotter()
mesh = ra.ctp()
streamlines = mesh.streamlines(vectors="e_l", source_radius=50, n_points=5000)
tubes = streamlines.tube()
plotter.add_mesh(mesh, opacity=0.5, color="white", label="myocardium")
plotter.add_mesh(tubes, color="red", label="fibers")
plotter.add_mesh(
pv.PolyData(right_atrium_appendage_coordinates),
color="blue",
point_size=20,
render_points_as_spheres=True,
label="right atrium appendage",
)
plotter.add_legend()
plotter.show()
C:\Users\ansys\actions-runner\_work\pyansys-heart\pyansys-heart\.tox\doc-html\Lib\site-packages\pyvista\core\utilities\points.py:77: UserWarning: Points is not a float type. This can cause issues when transforming or applying filters. Casting to ``np.float32``. Disable this by passing ``force_float=False``.
warnings.warn(
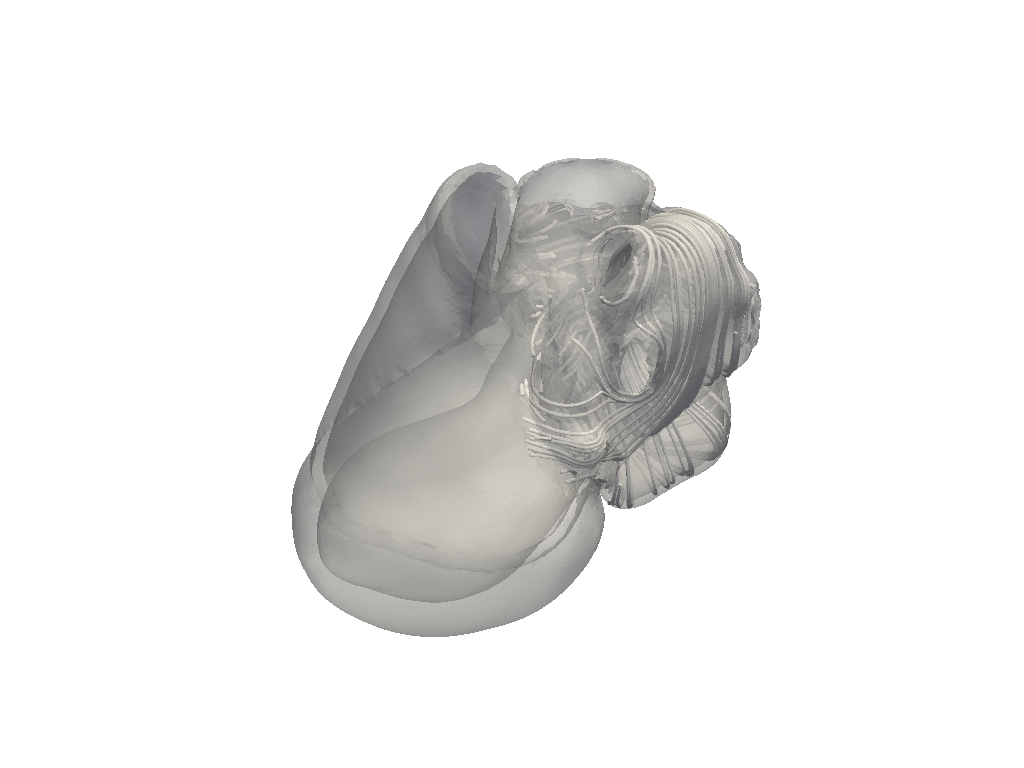
Plot full fibers on full heart model#
fiber_plot = model.plot_fibers(n_seed_points=5000)
Total running time of the script: (2 minutes 3.413 seconds)

