Note
Go to the end to download the full example code.
Define materials#
This example shows how to create a mechanical material and assign it to a heart part.
Import material module#
from pathlib import Path
import matplotlib.pyplot as plt
import ansys.health.heart.settings.material.cell_models as cell_models
from ansys.health.heart.settings.material.curve import kumaraswamy_active
import ansys.health.heart.settings.material.ep_material as ep_materials
from ansys.health.heart.settings.material.material import (
ACTIVE,
ANISO,
ISO,
ActiveCurve,
ActiveModel1,
ActiveModel3,
HGOFiber,
Mat295,
NeoHookean,
constant_ca2,
)
Note
The unit system for heart modeling in LS-DYNA is ["MPa", "mm", "N", "ms", "g"].
Create a material#
Create a Neo-Hookean material as follows.
neo = NeoHookean(rho=0.001, c10=1, nu=0.499)
C:\Users\ansys\actions-runner\_work\pyansys-heart\pyansys-heart\examples\preprocessor\demo-material_pr.py:62: DeprecationWarning: Call to deprecated class NeoHookean. (Use *MAT_295 with the ISO module instead.)
neo = NeoHookean(rho=0.001, c10=1, nu=0.499)
C:\Users\ansys\actions-runner\_work\pyansys-heart\pyansys-heart\examples\preprocessor\demo-material_pr.py:62: UserWarning: A custom validator is returning a value other than `self`.
Returning anything other than `self` from a top level model validator isn't supported when validating via `__init__`.
See the `model_validator` docs (https://docs.pydantic.dev/latest/concepts/validators/#model-validators) for more details.
neo = NeoHookean(rho=0.001, c10=1, nu=0.499)
The recommended way to create a Neo-Hookean material is by activating only the isotropic module in MAT_295.
neo2 = Mat295(rho=0.001, iso=ISO(itype=1, beta=2, kappa=1, mu1=0.05, alpha1=2))
Note
For more information on MAT_295, see the LS-DYNA manuals.
# Additional steps follow for creating MAT_295, which is used for myocardium.
# step 1: create an isotropic module
iso = ISO(k1=1, k2=1, kappa=100)
# step 2: create an anisotropic module
fiber = HGOFiber(k1=1, k2=1)
aniso1 = ANISO(fibers=[fiber])
# Create fiber with sheet, and their interactions
sheet = HGOFiber(k1=1, k2=1)
aniso2 = ANISO(fibers=[fiber, sheet], k1fs=1, k2fs=1)
# step3: create the active module
# example 1:
# create active model 1
ac_model1 = ActiveModel1()
# create Ca2+ curve
ac_curve1 = ActiveCurve(
func=constant_ca2(tb=800, ca2ionm=ac_model1.ca2ionm), type="ca2", threshold=0.5
)
# build active module
active = ACTIVE(model=ac_model1, ca2_curve=ac_curve1)
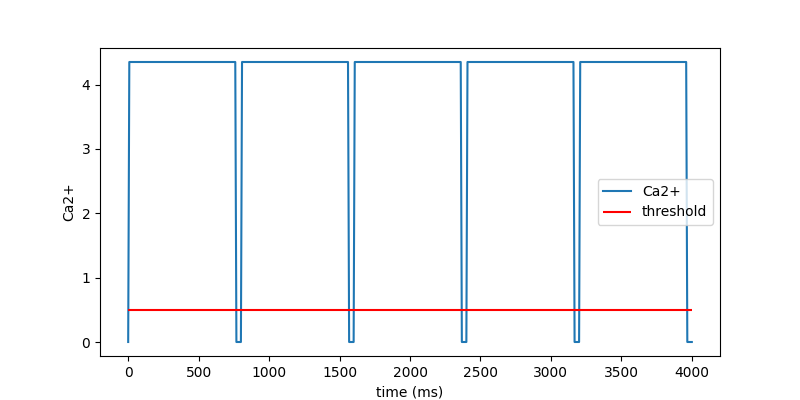
## Active model 1 must have a constant ca2ion,
# but the curve must cross the threshold at every start of the heart beat.
Plot Ca2+ with threshold#
Plot Ca2+ with the threshold.
fig = active.ca2_curve.plot_time_vs_ca2()
plt.show()
# example 2
# create active model 3
ac_model3 = ActiveModel3()
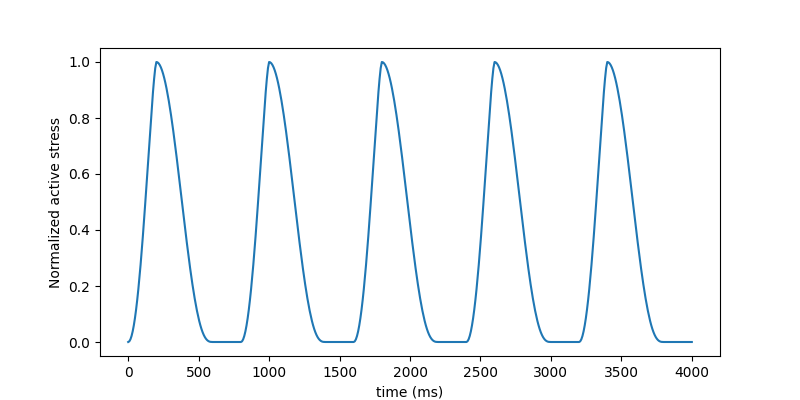
# create a stress curve and show
ac_curve3 = ActiveCurve(func=kumaraswamy_active(t_end=800), type="stress")
fig = ac_curve3.plot_time_vs_stress()
plt.show()
Note
When eta=0 in model 3, the stress curve is the active stress for all elements. If eta!=0, this is the idealized active stress when fiber stretch stays at 1.
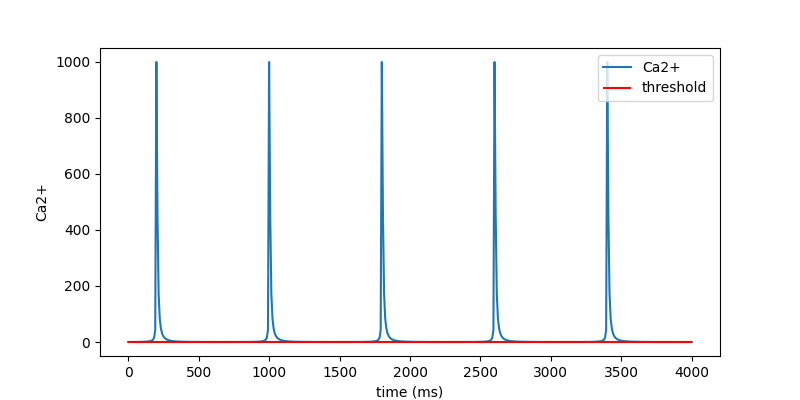
# PyAnsys Heart converts the stress curve to Ca2+ curve (input of MAT_295)
fig = ac_curve3.plot_time_vs_ca2()
plt.show()
# build active module
active3 = ACTIVE(model=ac_model3, ca2_curve=ac_curve3)
Create MAT_295 with modules#
Create MAT_295 with the preceding modules.
iso_mat = Mat295(rho=1, iso=iso, aniso=None, active=None)
passive_mat = Mat295(rho=1, iso=iso, aniso=aniso1, active=None)
active_mat = Mat295(rho=1, iso=iso, aniso=aniso1, active=active)
Create EP materials#
ep_mat_active = ep_materials.Active(
sigma_fiber=1, sigma_sheet=0.5, beta=140, cm=0.01, cell_model=cell_models.Tentusscher()
)
epinsulator = ep_materials.Insulator()
# import pyvista as pv
# ##############################################################################
# .. note::
# The Ca2+ curve is ignored if the simulation is coupled with electrophysiology.
Assign materials to a part#
Assign the materials to the heart model.
Load a heart model#
Note
You must complete the full heart preprocessing example first.
import ansys.health.heart.examples as examples
import ansys.health.heart.models as models
heart_model_vtu, heart_model_partinfo, _ = examples.get_preprocessed_fullheart()
workdir = str(Path.home() / "pyansys-heart" / "Rodero2021")
# Load a full-heart model.
heartmodel: models.FullHeart = models.FullHeart.load_model(
heart_model_vtu, heart_model_partinfo, working_directory=workdir
)
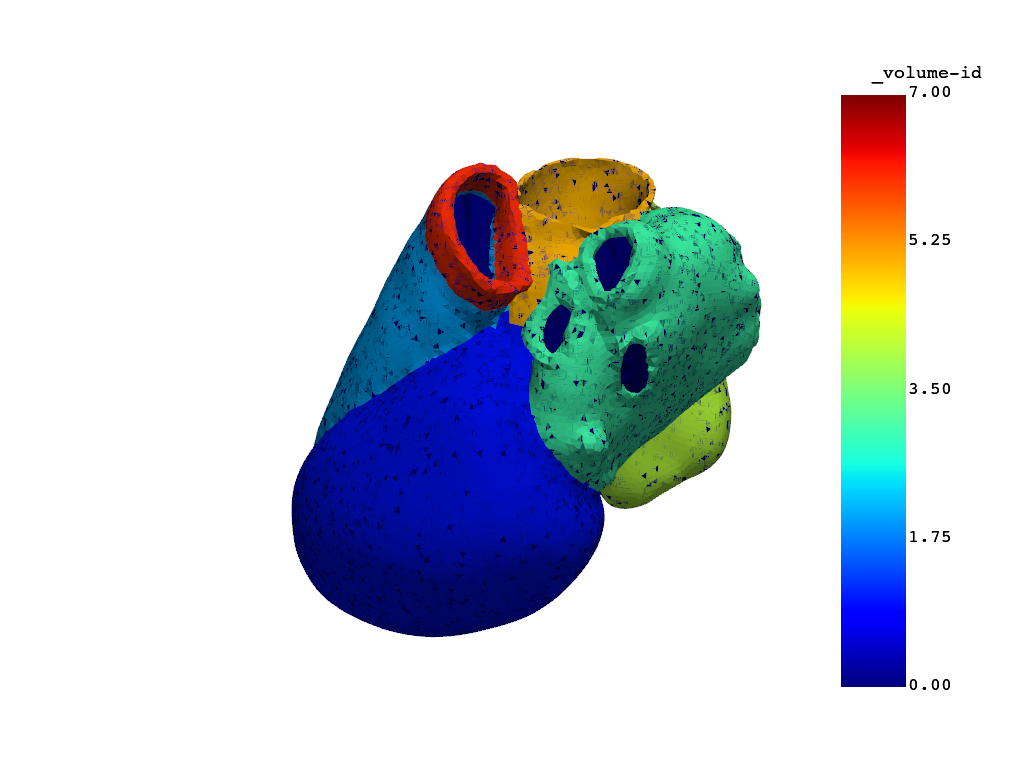
heartmodel.plot_mesh(show_edges=False)
# Print the default material. You should see that the material is empty.
print(heartmodel.left_ventricle.meca_material)
print(heartmodel.left_ventricle.ep_material)
None
None
Note
If no material is set before writing k files, the default material
from the settings object is used.
# Assign the material that you just created.
heartmodel.left_ventricle.meca_material = active_mat
heartmodel.left_ventricle.ep_material = ep_mat_active
# Print it. You should see the following:
# MAT295(rho=1, iso=ISO(itype=-3, beta=0.0, nu=0.499, k1=1, k2=1), aopt=2.0, aniso=ANISO(atype=-1, fibers=[HGOFiber(k1=1, k2=1, a=0.0, b=1.0, _theta=0.0, _ftype=1, _fcid=0)], k1fs=None, k2fs=None, vec_a=(1.0, 0.0, 0.0), vec_d=(0.0, 1.0, 0.0), nf=1, intype=0), active=ActiveModel.Model1(t0=None, ca2ion=None, ca2ionm=4.35, n=2, taumax=0.125, stf=0.0, b=4.75, l0=1.58, l=1.85, dtmax=150, mr=1048.9, tr=-1629.0)) # noqa
print(heartmodel.left_ventricle.meca_material)
print(heartmodel.left_ventricle.ep_material)
rho=1.0 iso=ISO(itype=-3, beta=0.0, nu=0.4950166112956811, k1=1.0, k2=1.0, mu1=None, alpha1=None, kappa=100.0) aopt=2.0 aniso=ANISO(atype=-1, fibers=[HGOFiber(k1=1.0, k2=1.0, a=0.0, b=1.0)], k1fs=None, k2fs=None, vec_a=(1.0, 0.0, 0.0), vec_d=(0.0, 1.0, 0.0), nf=1, intype=0) active=ACTIVE(acid=None, actype=1, acthr=0.5, acdir=1, sf=1.0, ss=0.0, sn=0.0, model=ActiveModel1(t0=None, ca2ion=None, ca2ionm=4.35, n=2, taumax=0.125, stf=0.0, b=4.75, l0=1.58, l=1.85, dtmax=150, mr=1048.9, tr=-1629.0), ca2_curve=ActiveCurve(func=(array([ 0., 8., 16., 24., 32., 40., 48., 56., 64., 72., 80.,
88., 96., 104., 112., 120., 128., 136., 144., 152., 160., 168.,
176., 184., 192., 200., 208., 216., 224., 232., 240., 248., 256.,
264., 272., 280., 288., 296., 304., 312., 320., 328., 336., 344.,
352., 360., 368., 376., 384., 392., 400., 408., 416., 424., 432.,
440., 448., 456., 464., 472., 480., 488., 496., 504., 512., 520.,
528., 536., 544., 552., 560., 568., 576., 584., 592., 600., 608.,
616., 624., 632., 640., 648., 656., 664., 672., 680., 688., 696.,
704., 712., 720., 728., 736., 744., 752., 760., 768., 776., 784.,
792., 800.]), array([0. , 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35,
4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35,
4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35,
4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35,
4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35,
4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35,
4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35,
4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35,
4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 0. , 0. , 0. ,
0. , 0. ])), type='ca2', threshold=0.5, n_beat=5, time=array([ 0., 8., 16., 24., 32., 40., 48., 56., 64., 72., 80.,
88., 96., 104., 112., 120., 128., 136., 144., 152., 160., 168.,
176., 184., 192., 200., 208., 216., 224., 232., 240., 248., 256.,
264., 272., 280., 288., 296., 304., 312., 320., 328., 336., 344.,
352., 360., 368., 376., 384., 392., 400., 408., 416., 424., 432.,
440., 448., 456., 464., 472., 480., 488., 496., 504., 512., 520.,
528., 536., 544., 552., 560., 568., 576., 584., 592., 600., 608.,
616., 624., 632., 640., 648., 656., 664., 672., 680., 688., 696.,
704., 712., 720., 728., 736., 744., 752., 760., 768., 776., 784.,
792., 800.]), t_beat=800.0, ca2=array([0. , 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35,
4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35,
4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35,
4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35,
4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35,
4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35,
4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35,
4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35,
4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 4.35, 0. , 0. , 0. ,
0. , 0. ]), stress=None))
sigma_fiber=1.0 sigma_sheet=0.5 sigma_sheet_normal=0.5 beta=140.0 cm=0.01 lambda_=None cell_model=Tentusscher(gas_constant=8314.472, t=310, faraday_constant=96485.3415, cm=0.185, vc=0.016404, vsr=0.001094, vss=5.468e-05, pkna=0.03, ko=5.4, nao=140.0, cao=2.0, gk1=5.405, gkr=0.153, gna=14.838, gbna=0.0002, gcal=3.98e-05, gbca=0.000592, gpca=0.1238, gpk=0.0146, pnak=2.724, km=1.0, kmna=40.0, knaca=1000.0, ksat=0.1, alpha=2.5, gamma=0.35, kmca=1.38, kmnai=87.5, kpca=0.0005, k1=0.15, k2=0.045, k3=0.06, k4=0.005, ec=1.5, maxsr=2.5, minsr=1.0, vrel=0.102, vleak=0.00036, vxfer=0.0038, vmaxup=0.006375, kup=0.00025, bufc=0.2, kbufc=0.001, bufsr=10.0, kbufsf=0.3, bufss=0.4, kbufss=0.00025, gks=0.392, gto=0.294, v=-85.23, ki=136.89, nai=8.604, cai=0.000126, cass=0.00036, casr=3.64, rpri=0.9073, xr1=0.00621, xr2=0.4712, xs=0.0095, m=0.00172, h=0.7444, j=0.7045, d=3.373e-05, f=0.7888, f2=0.9755, fcass=0.9953, s=0.999998, r=2.42e-08)
Create a new part and set material
# # A new part can be created by elements IDs
# ids = np.where(heartmodel.mesh.point_data_to_cell_data()["uvc_longitudinal"] > 0.9)[0]
# new_part: Part = heartmodel.create_part_by_ids(ids, "new_part")
# # Show the part
# plotter = heartmodel.plot_part(new_part)
Total running time of the script: (0 minutes 7.235 seconds)