Note
Go to the end to download the full example code.
Run a full-heart electrophysiology simulation#
This example shows how to consume a full-heart model and set it up for the main electrophysiology simulation. It loads a pre-computed heart model and computes the fiber orientation, Purkinje network, and conduction system. It then simulates the electrophysiology.
Warning
When using a standalone version of the DPF Server, you must accept the license terms. To accept these terms, you can set this environment variable:
import os
os.environ["ANSYS_DPF_ACCEPT_LA"] = "Y"
Perform the required imports#
Import the required modules and set relevant paths, including that of the working directory, heart model, and LS-DYNA executable file.
import os
from pathlib import Path
from ansys.health.heart.examples import get_preprocessed_fullheart
import ansys.health.heart.models as models
from ansys.health.heart.settings.settings import FibersDRBM
from ansys.health.heart.simulator import DynaSettings, EPSimulator
# Set the working directory and path to the model. This example assumes that there is a
workdir = Path.home() / "pyansys-heart" / "downloads" / "Rodero2021" / "01" / "FullHeart"
path_to_model, path_to_partinfo, _ = get_preprocessed_fullheart(resolution="2.0mm")
Load the full-heart model#
Load the full-heart model.
model: models.FullHeart = models.HeartModel.load_model(
path_to_model, path_to_partinfo, working_directory=workdir
)
# Save the model.
model.mesh.save(os.path.join(model.workdir, "simulation_model.vtu"))
Instantiate the simulator#
Instantiate the simulator and define settings.
Note
The DynaSettings object supports several LS-DYNA versions and platforms,
including smp, intempi, msmpi, windows, linux, and wsl.
Choose the one that works for your setup.
# Specify the LS-DYNA path. (The last tested working version is ``intelmpi-linux-DEV-106117``.)
lsdyna_path = r"ls-dyna_msmpi.exe"
# Instantiate LS-DYNA settings.
dyna_settings = DynaSettings(
lsdyna_path=lsdyna_path, dynatype="intelmpi", num_cpus=4, platform="windows"
)
# Instantiate the simulator, modifying options as necessary.
simulator = EPSimulator(
model=model,
dyna_settings=dyna_settings,
simulation_directory=os.path.join(workdir, "simulation-EP"),
)
Load simulation settings#
Load the default settings.
simulator.settings.load_defaults()
simulator.settings.electrophysiology.analysis.solvertype = "ReactionEikonal"
Compute fiber orientation#
Compute fiber orientation and plot the fibers on the entire model.
# Import the appendage landmarks.
from ansys.health.heart.pre.database_utils import right_atrium_appendage_landmarks
# Get the right atrium appendage landmark of the first case of Rodero2021.
right_atrium_appendage_coordinates = right_atrium_appendage_landmarks.get("Rodero2021").get(1)
# Initialize the D-RBM fiber settings.
d_rbm_settings = FibersDRBM()
print(d_rbm_settings)
# Compute ventricular fibers using the D-RBM method.
simulator.compute_fibers(fiber_settings=d_rbm_settings)
# Compute atrial fibers.
simulator.model.right_atrium.active = True
simulator.model.left_atrium.active = True
simulator.model.right_atrium.fiber = True
simulator.model.left_atrium.fiber = True
simulator.compute_left_atrial_fiber()
simulator.compute_right_atrial_fiber(appendage=right_atrium_appendage_coordinates)
simulator.model.plot_fibers(n_seed_points=1000)
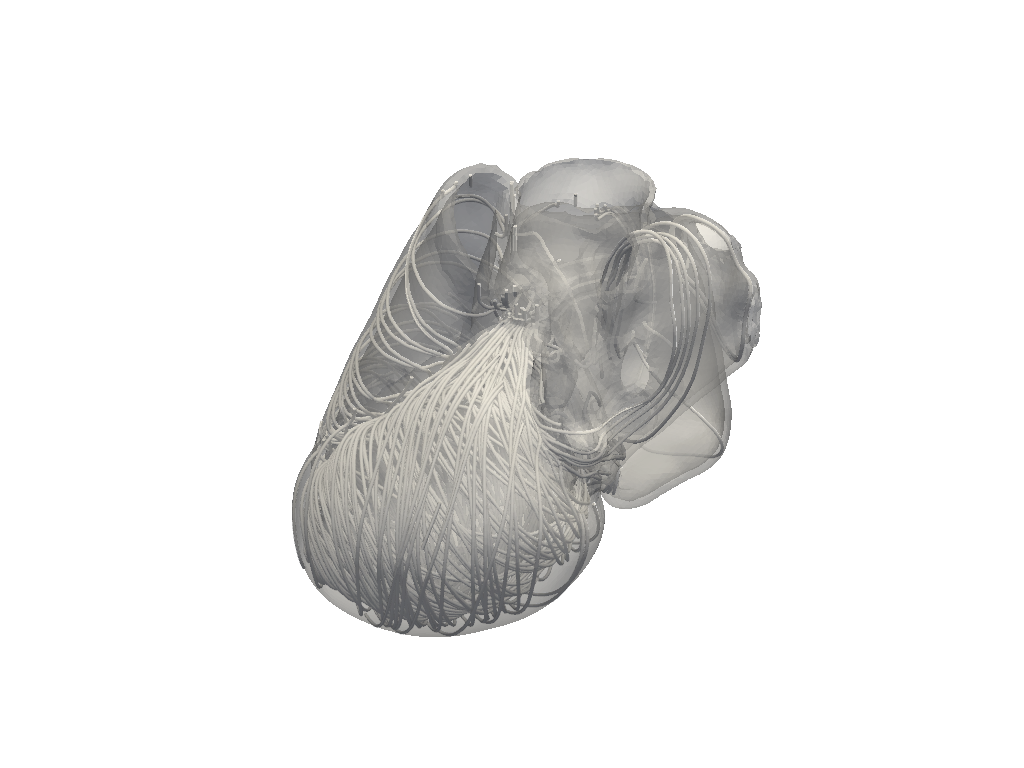
left_ventricle=BaseFiberSettings:
alpha_endo: 60 degree
alpha_epi: -60 degree
beta_endo: -20 degree
beta_epi: 20 degree
right_ventricle=BaseFiberSettings:
alpha_endo: -90 degree
alpha_epi: 25 degree
beta_endo: 0 degree
beta_epi: 20 degree
alpha_outflow_tract=None beta_outflow_tract=None septal_fraction=0.6666666666666666
<pyvista.plotting.plotter.Plotter object at 0x000002DFD18450D0>
Compute the conduction system#
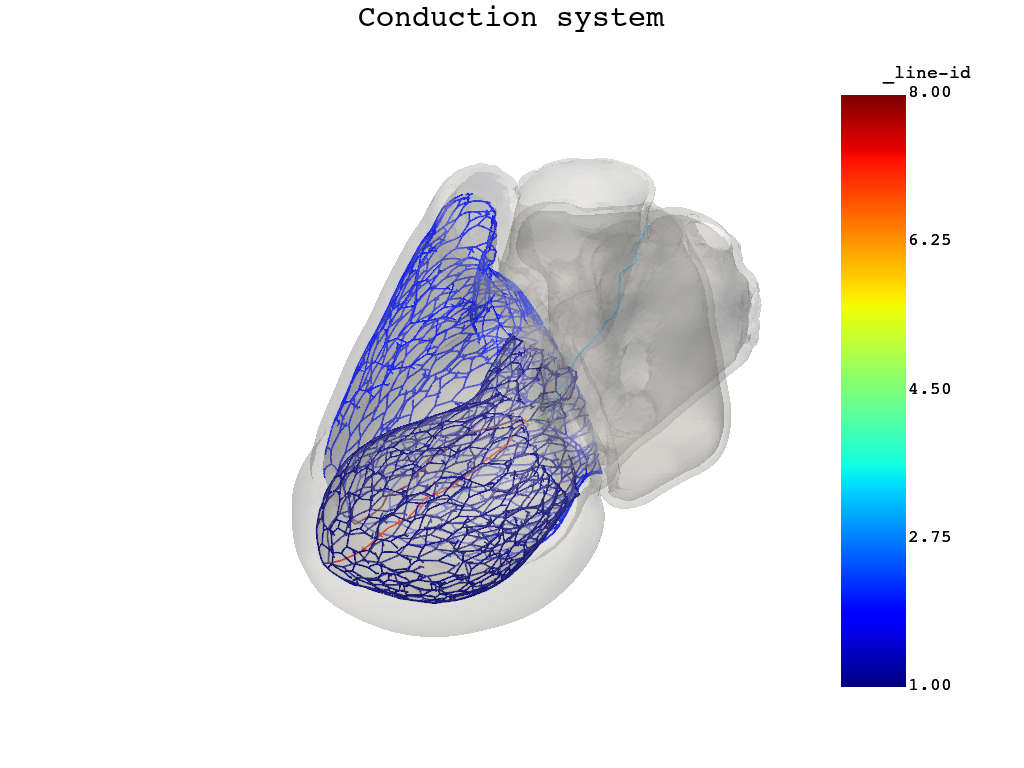
Compute the conduction system and Purkinje network, and then visualize the results. The action potential propagates faster through this system compared to the rest of the model.
simulator.compute_purkinje()
# By calling this method, stimulation is at the atrioventricular node.
# If you do not call this method, the two apex regions of the ventricles are stimulated.
simulator.compute_conduction_system()
simulator.model.plot_purkinje()
Start the main simulation#
Start the main electrophysiology simulation. This uses the previously computed fiber orientation and Purkinje network to set up and run the LS-DYNA model.
# Compute the Eikonal solution. This only computes the activation time.
simulator.simulate(folder_name="main-ep-ReactionEikonal")
Total running time of the script: (7 minutes 31.954 seconds)

